Shipboard CTD processing recipe#
Note
This is meant as a concrete, hands-on guide to CTD post-processing using NPI tools (including python libraries and a git template). The instructions are fairly new, so there will likely be things that are unclear, misleading, or wrong. Take from it what you find useful - and if you think somethign in these instructions can be made clearer, it is very useful if you can Øyvind F know.
This is a list of steps for post-processing shipboard CTD profile data. The goal of the processing is to go from raw data (.hex, .XMLCON, …) to a well-structured, quality-controlled, ready-for-analysis .nc file published FAIRly at the Norwegian Polar Data Centre. You may find some of the steps useful even if you have different goals (e.g. just processing without publishing).
While this list covers the most common steps, almost every dataset presents some unique issue that must be addressed individually. Therefore, it is important that the processor actively engages with the data and has a reasonable understanding of the workflow.
The list is very granular by design - some of these steps will likely be obvious if you have previous experience.
The list below outlines one possible approach using Python together with NPI’s kval library, which provides a range of specialized tools for this purpose.
1: Make a plan
Briefly summarize the context of data collection:
Where and when was the cruise?
Who participated in CTD work?
Were there any issues that could be useful to know about?
Were auxiliary data collected and processed (salinity samples, in-situ Chlorophyll-A, etc.)?
Outline a preliminary plan for data processing:
Who will perform the processing?
What are the main processing steps?
Will all variables meet the same quality/validation standards?
E.g., will uncalibrated data be included?
When will the processing be completed?
Plan the publication:
Who will publish the dataset?
When and where will this be published?
Who will be listed as authors?
Thinking through these points will help guide the process and be helpful in creating metadata later on.
2. Install necessary software
SBEDataProcessing — Seabird’s software package
Used in the initial data conversion/batch processing (#6).
Download from SBEDataProcessing - (Windows only).
Python — programming language.
There are many ways to set this up - see this for a Windows example.
It is recommended to use
condafor package management (the instructions in the guide above cover this).
3. Clone the template processing repository and create your own repo
Setup: git — version control software
Git is used for version control. You should carry out the processing in a directory on your laptop that corresponds to a repository on gitlab.npolar.no. This ensures proper documentation, reproducibility, and makes collaboration easier.
There are many ways to install and use git. The easiest approach here is to install it in your conda-enabled terminal (e.g. Miniforge for Windows users):
conda install -c conda-forge git
Setup: NPI Gitlab credentials
You should also ensure that you have a user with the necessary credentials at gitlab.npolar.no. Use your NPI credentials to sign in (the same username/password used to sign in on an NPI Windows laptop). To check that you have the necessary credentials, navigate to gitlab.npolar.no/npiocean/. You should see a Cruises category; if you don’t, get in touch with Øyvind and Tore with your gitlab.npolar.no username and they will grant you the necessary permissions.
Note you will need to be inside the NPI network to access and interact with
gitlab.npolar.no.
Create a new gitlab.npolar.no repository
We will create a repository in the NPI Gitlab instance. This will be the “central hub” for the work and also act as a backup and lasting documentation for the processing. You will have your own copy of this repository on your local computer, and sync this with the gitlab.npolar.no. You and other users can push and pull changes/updates, and these will be tracked, providing a complete records of the work that has been done.
Navigate to
gitlab.npolar.no/npiocean/cruisesin a browser.Select a subgroup (e.g.
TrollTransect,ArcticOcean,Other..)Alternatively, you can create a New subgroup if there’s not one that fits.
Click New project within this subgroup.
Select Create blank project.
Set a name (e.g.
mycruise_2022_ctdproc).Set Visibility level to
Internal.Toggle Initialize repository with a README to
Off.
You now have an (empty) repository for your CTD processing work. It should have a Git remote URL on the form:
git@gitlab.npolar.no:npiocean/cruises/<your-subgroup>/<your-repo>.git(SSH)Easiest once set up (no passsord authentication) but requires your to register an SSH key at gitlab.npolar.no.
https://gitlab.npolar.no/npiocean/cruises/<your-subgroup>/<your-repo>.git(HTTPS)Requires password authentication every time you interact with the remote repo.
Click
Codefrom your gitlab.npolar.no repository front page to display the Git URL.
To change the project name, enter a project description, set a repository avatar image etc., go to
Settings-Generalin the left sidebar on your gitlab.npolar.no repository page (optional).
Clone the CTD template to your system
This step copies a predefined template (directory structure, some template documents and scripts) to your system.
Clone the
ctd-processing-templaterepository to your computer:cd path/where/i/will/be/working/ % Clone via SSH if you have ssh key enabled: git clone git@gitlab.npolar.no:npiocean/processing-templates/ctd-processing-template.git % .. or via HTTPS if you don't (will require user/password authentication): git clone https://gitlab.npolar.no/npiocean/processing-templates/ctd-processing-template.git
This will create a copy of the template repository on your computer at
path/where/i/will/be/working/.Rename the directory to a title describing your dataset, e.g.
mycruise_2026_ctdproc.
Initialize a new repository
Modify the header of the file
README.mdfrom# Processing shipboard CTD from *{Cruise NN}*for your specific cruise.Initialize the directory as a new git repository, and stage and commit the change you just made:
cd {path/where/i/will/be/working/mycruise_2026_ctdproc} rm -rf .git % Remove connection to template repository git init % Initialize a new repository git add . % Stage any changes you've made in the repo git commit -m 'Initial commit' % Commit your staged changes with a (mandatory) description of the changes you have made
We now need connect your local processing folder (e.g. mycruise_2022_ctdproc/) from the previous step.
Sync local and remote repos
We will now connect the local directory on your computer to the remote repository on
gitlab.npolar.noso that we can push and pull changes between local and remote repositories.cd mycruise_2022_ctdproc git remote add origin [Git URL] % SSH or HTML, see above. git branch -M master git push -uf origin master
If you now navigate to the front page of your gitlab.npolar.no repository, you should see the commit you just made visible near the top of the page.
You can now
git add .->git commit -m '[comment]->git pushfrom your local computer to propagate local changes to the remote repositoryYou can also
git pullto propagate changes from the remote to the local, e.g. if a collaborator has made changes.It is a good idea to
git pushoften, and to alwaysgit pullbefore starting a new work session to avoid different versions diverging.
4. Populate your repository with source files
You should now have a working local repository - a directory structure with folders such as code, data/final, docs, etc, associated with a remote repository on gitlab.npolar.no. We will now populate the directory structure with files to prepare post-processing.
Grab source data files
Raw CTD files (
.hex,.XMLCON/.CON,.hdr,.bl..)Put these files in
data/source/ctd/raw/.
Cruise water sample log (excel sheet)
data/source/cruise_log/.
Salinometer log sheets
Scanned handwritten logsheets:
data/source/cruise_log/hand_written_logsheets.Logsheets digitized to Excel:
data/source/cruise_log/hand_written_logsheets.
Grab CTD configuration file
If your CTD configuration was identical for all stations - i.e. if no sensors or calibrations changed:
Copy one of the
.XMLCONfiles fromdata/source/ctd/raw/intosbe_processing/.Rename this file to
CTD_CONF.XMLCON.Alternatively, change
CTD_CONF.XMLCONto the name of your.XMLCONfile insbe_processing/scripts/batch_proc_all_template.bat.
If you use multiple configurations, you should have one .XMLCON file per configuration in sbe_processing/, and one sbe_processing/scripts/{}.bat script per configuration (each .bat script referring to one .XMLCON file).
5. Fill out basic information in README.md
Replace the dummy information in README.md with real information about the cruise and processing. Fill out what you can and write anything that could be useful - and feel free to edit and shape it however you want!
This file can be useful as an introduction to what is going in in the processing repository - for someone trying to reproduce your steps for another dataset, for someone interested in granular details about the data, or for yourself returning to the work after a few months..
6. Prepare a global metadata file
Locate the file
metadata/CTD_global_metadata.yaml.Edit this file to accurately reflect your cruise.
Any fields labelled
TBWmust be filled in.You can usually leave standard parameters as they are.
Later on, we will build metadata in out NetCDF file based on information in this file. Note that you can always go back and edit this .yaml file - but try to get as much right as you can at this stage.
There is also a file metadata/CTD_global_metadata.yaml, which by default is empty of content that would be imported. You may want to enter variable-specific metadata here.
7. Set up a dataset landing page at data.npolar.no
Open a browser and navigate to
data.npolar.no.Log in (
Log inin the bottom left corner, use your NPI e-mail and password).Click
New datasetin the upper rightFill out the Title and Summary fields
In many cases you can copy information directly from your
README.md.Summary should describe the dataset (not a project/cruise itself).
Note that you can use markdown, i.e. use subheadings, bullet lists, images etc.
Do what you can now - you can always revise later!
Fill out whatever other information you have available.
Keywords, geographical coverage, etc. Good to fill out what you can, but you can lways modify and add things.
Give collaborators editing permission:
Useful if you want to let others be able to edit the dataset page.
From the dataset editing page, click
Close.Click
⋮in the top corner, thenManage permissionsYou can now click
Addto give people read/edit permission.Note that this only works out of the box for collaborators at NPI who have logged tin at data.npolar.no at least once.
It is also possible, if sometwhat clunky, to add external collaborators. To do this, they have to log in at data.npolar.no. You then have to contant the Data Section to add your collaborator as a user.
Grab your DOI
From the dataset editing page, click
Close.You will now see a DOI lister under Other at the bottom of the page.
This dataset has been reserved for your dataset, and will be assigned to the dataset once published.
Note down the DOI
In your
README.mdIn the
doifield of yourCTD_global_metadata.yaml
NOTE You can return to your dataset editing page by logging in to data.npolar.no, clicking your dataset title (which you can search for) and clicking Edit.
8. Run SBE batch processing (.hex, .xmlcon → .cnv, .btl, ..)
We will now set up and run batch processing of all raw CTD data using the SBEDataProcessing software.
The SBE processing in this case consists of:
Data conversion.
(Gentle) temporal filtering.
Loop editing (removing parts of the data where the rosette has moved up and down in “loops”).
Calculating derived variables including salinity from primary variables like C/T/P.
Averaging profiles into bins of 1 dbar.
Creating bottle summary (
.btl) files used for comparison with water samples colelcted from the rosette.Window filtering CDOM (if it is one of the variables we have collected) using three consecutive filters.
Feel free to skip the SBE processing step in its entirety if you already have .cnv and .btl files that you are happy with - just make sure to place your files in the appropriate subfolder of data/source/ctd/sbe_proc/.
Notes:
This step must be performed in Windows. You’ll need either a Windows PC or a Windows virtual machine/emulator (e.g. Wine). If you are switching systems, you should make a clone of the gitlab repository to your windows machine where you’ll do the SBE processing, and
git pushyour changes when you are done.This process can be a little cumbersome, but it not quite as bad as it looks here (everything is just described in repetitive detail). The good news is that you should normally only have to do this once per cruise.
The steps below are based on the SBE Processing steps described in the
CTD-classicprocedure developed by Yannick. The procedure below has been modified to fit the template folder structure, bit the processing steps should be exactly the same.The recipe assumes you have all your raw data in place. You may also want to do this during the cruise before you have all the CTD data from the cruise. That is completely fine - just run the batch processing (not the setup) again when you have all files in place in your
data/ctd/source/ctd/raw/directory.
Setup processing files
Open the SBE Data Processing program and click Run. You will see a list of processing operations available in the software. The CTD-classic procedure specifies a combination of operations and associated parameters we will apply as part of the automated processing. To set up the batch processing, we have to configure and run each step once, saving the configuration to an associated .psa file in sbe_processing/psa_cruise/.
Data conversion
Open SBEDataProcessing ➔
Run➔1. Data Conversion...Set
Program setup fileto{cruise_dir}\sbe_processing\psa_cruise\DatCnv.psa.Set
Instrument configuration fileto{cruise_dir}\sbe_processing\CTD_CONF.XMLCON.Set output variables:
Under the
Data Setuptab, clickSelect output variables.Select all variables that were measured by the CTD on your cruise.
Remember to include
Latitude [deg],Longitude [deg],Time NMEA [seconds], if available.For dual sensors include both, e.g. both
TemperatureandTemperature, 2.Note: Do not include salinity here - we will compute salinity at a later stage.
Close and go back to
File Setup
Set
Input filesto one of your.hexfiles, e.g.{cruise_dir}\data\ctd\source\ctd\raw\001.hex.Set
Output directoryto{cruise_dir}\data\ctd\source\ctd\sbe_proc\.Leave everything else as it is and click
Start Process.Progress bar should go from 0% to 100% and this should produce
.cnvand.rosfiles insbe_proc\.
Click
Exitand clickYeswhen asked whether to save changes.
Filter
Open SBEDataProcessing ➔
Run➔2. Filter...Set
Program setup fileto{cruise_dir}\sbe_processing\psa_cruise\Filter.psa.Set
Input filesto (e.g.){cruise_dir}\data\ctd\source\ctd\sbe_proc\001.cnv.Set
Output directoryto{cruise_dir}\data\ctd\source\ctd\sbe_proc\Leave everything else as it is and click
Start Process.Click
Yes(ok to overwrite).Progress bar should go from 0% to 100% and this should update the
.cnvfilesbe_proc\.
Click
Exitand clickYeswhen asked whether to save changes.
Cell Thermal mass
Open SBEDataProcessing ➔
Run➔4. Cell Thermal Mass...Set
Program setup fileto{cruise_dir}\sbe_processing\psa_cruise\CellTM.psa.Set
Input filesto (e.g.){cruise_dir}\data\ctd\source\ctd\sbe_proc\001.cnv.Set
Output directoryto{cruise_dir}\data\ctd\source\ctd\sbe_proc\Leave everything else as it is and click
Start Process.Click
Yes(ok to overwrite).Progress bar should go from 0% to 100% and this should update the
.cnvfilesbe_proc\.
Click
Exitand clickYeswhen asked whether to save changes.
Loop Edit
Open SBEDataProcessing ➔
Run➔5. Loop Edit...Set
Program setup fileto{cruise_dir}\sbe_processing\psa_cruise\LoopEdit.psa.Set
Input filesto (e.g.){cruise_dir}\data\ctd\source\ctd\sbe_proc\001.cnv.Set
Output directoryto{cruise_dir}\data\ctd\source\ctd\sbe_proc\Leave everything else as it is and click
Start Process.Click
Yes(ok to overwrite).Progress bar should go from 0% to 100% and this should update the
.cnvfilesbe_proc\.
Click
Exitand clickYeswhen asked whether to save changes.
Note: You may in some cases want to change the
Soak depthparameters at this stage. See “Adjusting the surface soak depth parameter” below.
Derive
Open SBEDataProcessing ➔
Run➔6. Derive...Set
Program setup fileto{cruise_dir}\sbe_processing\psa_cruise\Derive.psa.Set
Instrument configuration fileto{cruise_dir}\sbe_processing\CTD_CONF.XMLCON.Set
Input filesto (e.g.){cruise_dir}\data\ctd\source\ctd\sbe_proc\001.cnv.Set
Output directoryto{cruise_dir}\data\ctd\source\ctd\sbe_proc\Leave everything else as it is and click
Start Process.Click
Yes(ok to overwrite).Progress bar should go from 0% to 100% and this should update the
.cnvfilesbe_proc\.
Click
Exitand clickYeswhen asked whether to save changes.
Bin Average
Open SBEDataProcessing ➔
Run➔8. Bin Average...Set
Program setup fileto{cruise_dir}\sbe_processing\psa_cruise\BinAvg.psa.Set
Instrument configuration fileto{cruise_dir}\sbe_processing\CTD_CONF.XMLCON.Set
Input filesto (e.g.){cruise_dir}\data\ctd\source\ctd\sbe_proc\001.cnv.Set
Output directoryto{cruise_dir}\data\ctd\source\ctd\sbe_proc\Leave everything else as it is and click
Start Process.Click
Yes(ok to overwrite).Progress bar should go from 0% to 100% and this should create a
_bin.cnvfile insbe_proc\.
Click
Exitand clickYeswhen asked whether to save changes.
Bottle Summary
Open SBEDataProcessing ➔
Run➔9. Bottle Summary...Set
Program setup fileto{cruise_dir}\sbe_processing\psa_cruise\BottleSum.psa.Set
Instrument configuration fileto{cruise_dir}\sbe_processing\CTD_CONF.XMLCON.Set
Input filesto (e.g.){cruise_dir}\data\ctd\source\ctd\sbe_proc\001.ros(❗)Set
Output directoryto{cruise_dir}\data\ctd\source\ctd\sbe_proc\Leave everything else as it is and click
Start Process.Click
Yes(ok to overwrite).Progress bar should go from 0% to 100% and this should create a
.btlfile insbe_proc\.
Click
Exitand clickYeswhen asked whether to save changes.
Window Filter
(3 filter operations)
Note: Only apply this step if your CTD data include CDOM measurements (
Fluorescence, WET Labs CDOM).
Open SBEDataProcessing ➔
Run➔13. Window Filter...Set
Program setup fileto{cruise_dir}\sbe_processing\psa_cruise\W_filter_short.psa.Set
Input filesto (e.g.){cruise_dir}\data\ctd\source\ctd\sbe_proc\001.cnv*Set
Output directoryto{cruise_dir}\data\ctd\source\ctd\sbe_proc\Leave everything else as it is and click
Start Process.Click
Yes(ok to overwrite).Progress bar should go from 0% to 100% and this should update the
.cnvfilesbe_proc\.
Click
Exitand clickYeswhen asked whether to save changes.
Open SBEDataProcessing ➔
Run➔13. Window Filter...Set
Program setup fileto{cruise_dir}\sbe_processing\psa_cruise\W_filter_long.psa.Set
Input filesto (e.g.){cruise_dir}\data\ctd\source\ctd\sbe_proc\001_bin.cnv.Set
Output directoryto{cruise_dir}\data\ctd\source\ctd\sbe_proc\.Leave everything else as it is and click
Start Process.Click
Yes(ok to overwrite).Progress bar should go from 0% to 100% and this should update the
.cnvfilesbe_proc\.
Click
Exitand clickYeswhen asked whether to save changes.
Open SBEDataProcessing ➔
Run➔13. Window Filter...Set
Program setup fileto{cruise_dir}\sbe_processing\psa_cruise\W_final_smooth.psa.Set
Input filesto (e.g.){cruise_dir}\data\ctd\source\ctd\sbe_proc\001_bin.cnvSet
Output directoryto{cruise_dir}\data\ctd\source\ctd\sbe_proc\Leave everything else as it is and click
Start Process.Click
Yes(ok to overwrite).Progress bar should go from 0% to 100% and this should update the
.cnvfilesbe_proc\.In the batch processing, this stepwill produce a file
_bin_cdom.cnv
Click
Exitand clickYeswhen asked whether to save changes.
Setup batch processing
Create a copy of the file
{cruise_dir}\sbe_processing\scripts\batch_proc_all_template.bat➔{cruise_dir}\sbe_processing\scripts\batch_proc_all.bat
If your CTD data no not include CDOM measurements (Fluorescence, WET Labs CDOM):
Edit the file
{cruise_dir}\sbe_processing\scripts\batch_proc_all.bat(e.g. right click ➔Edit in Notepad)Around line 70 of the file, change
set do_run_window_filtering=1toset do_run_window_filtering=0.
Run batch processing
To run the batch processing, simply double click {cruise_dir}\sbe_processing\scripts\batch_proc_all.bat. This should run all the processing steps as specified in the .psa files on all your raw data, producing .cnvs and .btl files (and more) for each CTD profile. You should see SBE Data Processing Windows popping out as the script works through all steps for all profiles. The whole thing can take a few minutes - you can follow the progress in the terminal window.
NOTE: If something is set up wrong, you may experience that the terminal window that opens immediately closes with nothing else happening. To troubleshoot, it might help to run
batch_proc_all.batfrom the terminal:
Press
Win+R.Type
cmdand hit Enter.Navigate to the folder contain the script:
cd {cruise_dir}\sbe_processing\scripts\.Run the script by typing
batch_proc_all.bathitting Enter.
Adjusting the surface soak depth parameter
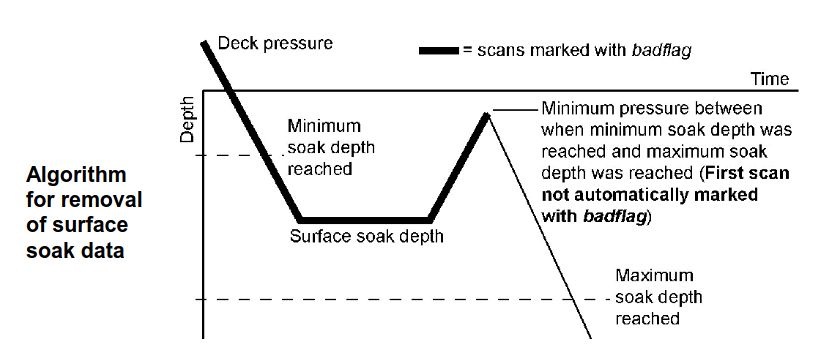
It is possible - and occasionally necessary - to modify any of the parameters involved in the SBE processing operations, or the combinations of operations themselves. This document doesn’t contain information about the details of the SBE processing (look for them in the Seasoft V2 Data Processing Software Manual), but one common modification is changing the surface soak depth parameters in the Loop Edit processing step. These parameters are used to identify and remove the part of the profile where the CTD is “soaked” at an initial depth, typically 20-50 m, before the profile begins. (Soaking is done to activate the conductivity pump -the CTD is subsequently brought up to right below the surface and then the actual downcast profile begins).
The
SurfaceSoakDepthMin/Minparameters inLoopEdit.psaspecify a pressure range within which the CTD is stopped to soak before being brough up again. Problems typically occur if the CTD was soaked at a depth outside this range. You may see this as messy data (especially conductivity/salinity) in the upper tens of meters as the profile includes parts of the cast where the conductivity pump was not turned on.
The solution to this is typically to:
Identify the actual soak depth (how deep did the CTD go before being pulled up to the surface to start the cast).
Modify the
SurfaceSoakDepthMaxandSurfaceSoakDepthMinparameters inLoopEdit.psa.
You can do this in a text editor, or in the SBE software - see page 103 of the SBE Processing Software Manual.
Note that there is also a parameter just called
SurfaceSoakDepth- this doesn’t seem to actually do anything.Rerun the SBE processing, producing updated
.cnvs.For example, if your
LoopEdit.psaspecifiedSurfaceSoakDepthMax=15andSurfaceSoakDepthMax=25, but the CTD for some reason was soaked at 30 m, you will very likely see junk in the top of your profile. You should make sense that theSurfaceSoakDepthMaxparameter is greater than 30 (something like 35 might be good), e.g.:
<SurfaceSoakDepthMin value="15.000000" />
<SurfaceSoakDepthMax value="25.00000035.000000" />
9. Setup your python environment and fire up JupyterLab
We now need to install the Python packages necessary to run the post-processing. This includes JupyterLab (a web-based interactive development environment for notebooks, code, and data) and NPI’s Python libraries kval (Python tools for processing and analysis of ocean data), and naust (NPI-specific tools and templates).
The easiest way to do this is to install a pre-made conda environment where everything has been set up for you. To do this, run this in your a conda-enabled terminal like Miniforge (from the root directory of your repository - where you should have a file ctd_env.yaml):
conda env create -f ctd_env.yaml
This installs a Python environment ctd_env with compatible versions of kval, naust, jupyter, and other required libraries.
To work within this environment, activate it in your conda terminal:
conda activate ctd_env
.. and while we’re at it: register the kernel in Jupyter (never mind what this means for now; also not strictly necessary) by pasting the following into your terminal
python -m ipykernel install --user --name ctd_env --display-name "CTD conda environment"
Alternative: Installing packages directly from conda
You can also install packages into an environment yourself. This usually works, but there is a larger risk that you may run into version conflicts etc. You should make sure your environment contains kval (version 0.4.3), naust (version 0.1.2) and jupyter.
To install kval:
Latest published version:
conda install -c npiocean -c conda-forge kval
Development version (if you want the newest features):
Clone the source repository and install in editable mode:
git clone https://github.com/NPIOcean/kval.git cd kval pip install -e .
(The
cd kvalcommand works in Miniforge/PowerShell and Linux/Mac shells; in >cmd.exeit may look slightly different.)The
-eflag installs the package in editable mode, meaning you can update the code (e.g., by pulling from GitHub) and your Python environment will use the updated version immediately.
You can now start up JupyterLab and begin working with notebooks:
First, activate the conda environment (or another environment with all requirements installed):
conda activate ctd_envNavigate to your root project folder, and start Jupyter Lab:
jupyter labThis should open a window in your browser. You can now browse in the folder structure using the left side menu, open existing notebooks or files, or create new ones.
When running a notebook, you can specify which Python kernel to use. For CTD processing, you should select the CTD environment we created earlier (or another environment with all the requirements installed):
Click
Kernel->Change kernel..in the top menu.Choose
CTD conda environment(this option should be available if you registered the kernel - enabled JupyterLab to see your conda environment - in #9)
10. Perform salinity QC
If salinity samples were collected during your cruise, laboratory salinity values should be compared with CTD-derived salinity values (from .btl files produced in Step #8). Salinity from water samples first have to be measured using a salinometer (see this).
If you have all the necessary inputs, you should be able to do this by opening JupyterLab (see #9) and running a few lines in
code/salt_qc/Salinity QC.ipynbMore detailed instructions here: QC of CTD salinity data
11. Calibrate oxygen/Chl-A/etc
To obtain high-quality CTD data on Chlorophyll-A, oxygen and other parameters, the CTD-derived values typically need to be calibrated against in-situ samples that have been processed in the lab (filtering and chl-a measurements, winkler titrations).
Typically, this amonts to something like a linear fit based of laboratory values against CTD measurements at bottle closure (.btl files). There are no detailed instructions for how to do this here.
The outcome of such a laboratory calibration is typically:
A mapping used to convert uncalibrated CTD values to calibrated values (e.g. \(x_2 = Ax_1 + b\)).
Will be applied to the data.
A description of how the calibration was done.
Will be applied to the dataset metadata.
In some cases, an additional correction is applied to compensate for non-photochemic quenching using transmissometer attenuation data (see Thomalla et al. 2017). In this case, you will have to format the metadata accordingly.
12. Inspect the data
Use JupyterLab to run the notebook code/ctd/1. Inspect CTD.ipynb.
Note: Make sure to run with an appropriate kernel. If you installed the conda environment
ctd_env.yamland registered the kernel in Step 2:Ckick
Kernel->Change kernel..in the top menu.Choose
CTD conda environment
Running this notebook doesn not modify anything, but should make sure data can be loaded and give you a good overview of what’s in the data.
In this notebook, we:
Try loading the data from
.cnvs to an xarray dataset.Plot a quick map of the CTD stations as a sanity check.
Produce dynamic contour plots of the CTD profile data - useful for getting the feel of what we are measuring.
Inspect individual profiles (you should visually inspect all profiles you intend to publish at least once!).
Plot variables agains each other (plot in “phase space”) - e.g. T-S diagrams - to dig a bit deeper into anything you find suspicious.
Compare dual sensors (e.g. dual conductivity sensors) to identify issues or offsets between the two.
For anything not covered by the statndard kval functions, you may want to create your own plots. Look up basic python/matplotlib syntax - it’s not too hard even if you’re not familiar.
It is good to make some notes in the notebook (at the bottom of the notebook for example), summarizing your impression and noting down any specific problems. This will be useful in the rest of the process.
13. Edit the data
Run the notebook code/ctd/2. Edit CTD.ipynb. In this step, we load the data from .cnv files to an xarray Dataset, perform some operations, and save to a temporary NetCDF file.
In this notebook:
Load the data from
.cnvs to an xarray dataset.Apply any corrections, e.g.:
Inspect disctribution of values and apply thresholds for valid data if necessary
Apply fixed offsets if that has been deemed necessary besed on other analysis
Apply (linear) chlorophyll calibration based on comparison between fluorometer and in-situ values.
Edit outliers
This is typically necessary at least for salinity, where we usually have some spikes due to particles in the conductivity cell.
The
hand_remove_points()function helps you edit out these points by hand.
Save to a temporary NetCDF which we will load in the final post-processing step.
You may want to also execute any other code used to modify data in this notebook (we will deal with metadata in the next one).
14. Finalize the file and metadata
Run the notebook code/ctd/3. Finalize CTD.ipynb. In this step, we load the NetCDF file produced in the previous step, and make changes to the file structure and metadata until we are happy with it and everything is compliant with CF/ADCC conventions.
In this notebook:
Load the data from
.ncproduced in step 12.Remove any variables we don’t want to include in the final dataset.
Import metadata attributes from
metadata/CTD_global_metadata.yamlandmetadata/CTD_variable_metadata.yaml(see Step 6).Automatically add some metadata based on the data (geospatial and temporal ranges, standard attributes, keywords etc).
Add a
date_createdattributebased on today’s date.Convert all 64-bit floats and ints to 32-bit - recommended for robustness across syetems.
Convert any NaNs to numerical fillvalues (default -9999) - recommended for robustness across syetems and to minimize ambiguity.
Run some checks:
Run the IOOS CF-convention checker
Run a custom, complementary checker in kval
Covers some common issues not covered by the IOOS checker
Note: These checkers are very useful, but they don’t catch everything. You should also be checking carefully yourself!
Produce a final, well-formatted NetCDF file.
In principle, this shoudl now be publication-ready, in practice you may want to return to update tweak things, e.g. after reaching out to coauthors or double checking.
Optionally also export a
.yamlfile mirroring the contents of the NetCDF, and a.matfile if you have collaborators that prefer that.What you shoudl publish is the
.ncfile.
In practice, you will probably want to work iteratively here. Make some changes, encounter something weird further down, go back and modify something, run a checker, modify somethign else flagged by the checker, etc.. When you pass the checkers without any major issues and you have looked thorugh the data and metadata yourself, you probably have a really well-formatted NetCDF.
At this stage, you may want to share your NetCDF, and the associated
.yamlfile, with collaborators to make sure everyone involved is happy with the file and metadata. You can always come back to this notebook and make changes.
14. Upload your files to data.npolar.no
Go to your data.npolar.no dataset landing page -> Edit -> Files. Here, you may Add individual files or Add directory to add an entire folder (useful for raw data).
Good to upload both the netCDF file produced in step 13 and a folder containing the source SBE files (e.g.:
source/rawfor.hex, .xmlconandsource/human_readable/for.cnv, .btl).Optional: also include the notebook/script to reproduce the processing
Note you can delete uploaded files and make other changes until the dataset is published. When the dataset is public, you can not remove or modify the uploaded files (however, you can upload newer files, e.g. _v2.nc).
15. Look over the dataset page
Are the correct files uploaded?
Is the landing page information and metadata information (time span, geographic area, dataset description, keywords, authors..) correct?
Are things set up such that you can easily upload a newer version if the data if you need to later?
Are the correct links (to cruise reports, projects etc) in place?
16. Make the dataset public
When you are happy with the data and metadata, and have double checked everything:
Go to your
data.npolar.nodataset landing pageClick
Publish, andOkto confirm.This will
Activate the dataset DOI
Make the dataset publicly visible (no login required).
Lock attached files in place (they can no longer be removed).
Hooray!

17 (for KPH): Communicate files to IMR
IMR will integrate processed KPH CTD data into Copernicus, data repositories used for generating ocean climatologies, etc. Notify them about the finalized dataset. (Contact Helge Sagen).
18. Clean-up
When you are done, you should:
Clean up the Gitlab repo (especially: undate
README.md).Your goal shoudl be to help someone else (or yourself in the future) understand what goes on in the repository.
Delete the
ctd_envconda environment:conda env remove -n ctd_env
Not necessary, but good practice. Also prevents any confusion next time you want to install this environment from
ctd_env.yaml(which may be updated later).
If anything in the process, scripts, or instructions felt confusing or clunky, just send a quick message to Øyvind. Feedback is super helpful and will help make this process easier next time someone does it.
Pat yourself on the back, and notify collaborators or any other interested parties that the dataset is publicly available.